The Pico Time-Digitizing Correlator Data Viewer { Windows Menu }

TG Graphing Window
Clicking the 'TG Graphing Window' menu item opens the
TG Graph:
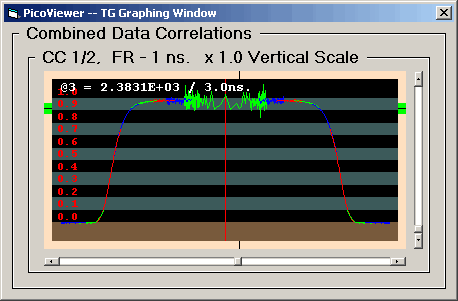
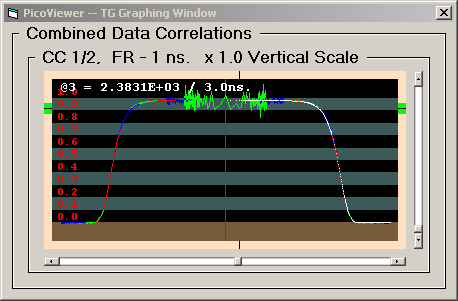
The TG Graph displays auto or cross correlation
spectra with '0' time at the center and larger times
extending toward the left and right edges. This graph
displays the data logarithmically in time with from
1 to 14 overlapping data sets. Each data set has a
time range of two decades:
1 - 100 100 ps bins (FC data)
or 100 50 ps bins (FC data)
or 100 33 ps bins (FC data)
2 - 100 1 ns bins (Rebinned FC data)
3 - 100 10 ns bins (Rebinned FC data)
4 - 100 100 ns bins (Rebinned FC data)
5 - 100 1 us bins (BN data)
6 - 100 10 us bins (BN data)
7 - 100 100 us bins (BN data)
8 - 100 1 ms bins (BN data)
9 - 100 10 ms bins (BN data)
10 - 100 100 ms bins (BN data)
11 - 100 1 s bins (BN data)
12 - 100 10 s bins (BN data)
13 - 100 100 s bins (BN data)
14 - 100 1000 s bins (BN data)
Each displayed data set has been normalized using
only the acquisition time and count rates to produce
the standard correlation display where a value of 1.0
is the baseline. The display baseline is at the top
of the brown stripe in the display. The alternate
black and gray stripes are .1 intervals. The maximum
correlation value of 2.0 is at the top of the upper
gray stripe.
The horizontal scroll bar allows one to move the
cross hairs (in the tan colored area around the spectra)
to a particular time. The channel number, counts in the
channel, and the correlation time are displayed at the
top of the spectra. The vertical scroll bar may be used
to change the vertical scaling. The current vertical
scaling factor is displayed above the spectra.
The 'Graphing Control' tab associated with the displayed
TG data is:
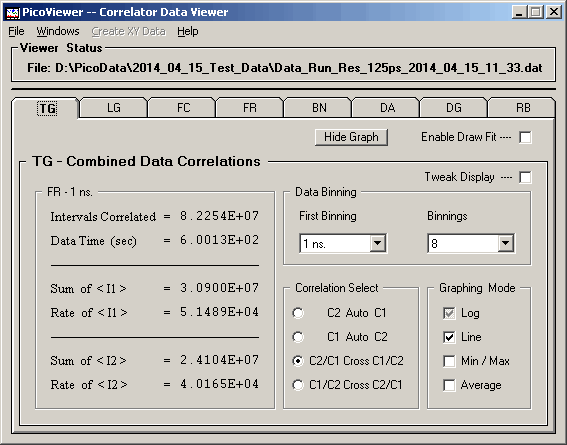
The TG tab options include the ability to select
the first displayed data set and the number of data sets
to display. The 'Average' option numerically averages
the left / right spectra and displays the average on
both the left and right sides of '0'.
The 'Tweak Display' check box enables the normalization
of the displayed FR spectra (.0625/.125, 1, 10, and 100ns
binnings) to the BN spectra (1 microsecond and larger) by
using a 3 microsecond interval starting at the 9 microsecond
point. The correlation method used for the FR spectra is
sensitive to large variations in the count rate during
acquisition. This results in a very small decrease in the
perceived count rate causing a small, usually less than
a few tenths of a percent, mismatch between the FR and
BN spectra.
The 'Enable Draw Fit' check box in conjunction with the
selection of a suitable equation and fitting parameters on
the MrqFit -- Data Fitting window allows the
display of a data fit superimposed upon the acquired data.

LG Graphing Window
Clicking the 'LG Graphing Window' menu item opens the
LG Graph:
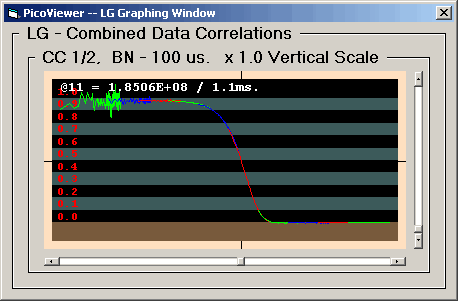
The LG Graph displays one of the correlation
spectra with '0' time at the left. This graph
displays the data logarithmically in time with from
1 to 14 overlapping data sets. Each data set has a
time range of two decades:
1 - 160 62.5 ps bins (FC data)
or 80 125 ps bins (FC data)
2 - 100 1 ns bins (Rebinned FC data)
3 - 100 10 ns bins (Rebinned FC data)
4 - 100 100 ns bins (Rebinned FC data)
5 - 100 1 us bins (BN data)
6 - 100 10 us bins (BN data)
7 - 100 100 us bins (BN data)
8 - 100 1 ms bins (BN data)
9 - 100 10 ms bins (BN data)
10 - 100 100 ms bins (BN data)
11 - 100 1 s bins (BN data)
12 - 100 10 s bins (BN data)
13 - 100 100 s bins (BN data)
14 - 100 1000 s bins (BN data)
Each displayed data set has been normalized using
only the acquisition time and count rates to produce
the standard correlation display where a value of 1.0
is the baseline. The display baseline is at the top
of the brown stripe in the display. The alternate
black and gray stripes are .1 intervals. The maximum
correlation value of 2.0 is at the top of the upper
gray stripe.
The horizontal scroll bar allows one to move the
cross hairs (in the tan colored area around the spectra)
to a particular time. The channel number, counts in the
channel, and the correlation time are displayed at the
top of the spectra.
The vertical scroll bar may be used to change the
vertical scaling. The current vertical scaling factor
is displayed above the spectra.
The 'Graphing Control' tab associated with the displayed
LG data is:
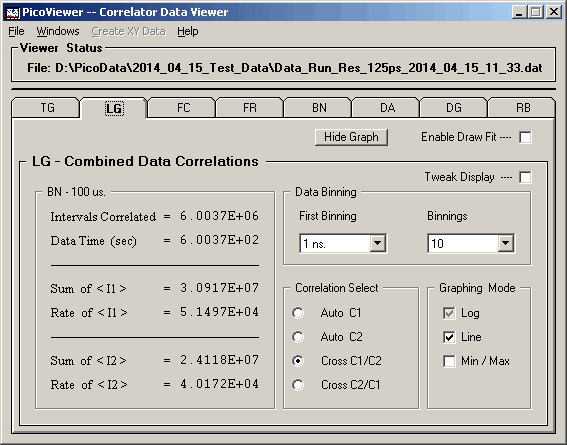
The LG tab options include the ability to select
the first displayed data set and the number of data sets
to display.
The 'Tweak Display' check box enables the normalization
of the displayed FR spectra (.0625/.125, 1, 10, and 100ns
binnings) to the BN spectra (1 microsecond and larger) byusing a
using a 3 microsecond interval starting at the 9 microsecond
point. The correlation method used for the FR spectra is
sensitive to large variations in the count rate during
acquisition. This results in a very small decrease in the
perceived count rate causing a small, usually less than
a few tenths of a percent, mismatch between the FR and
BN spectra.
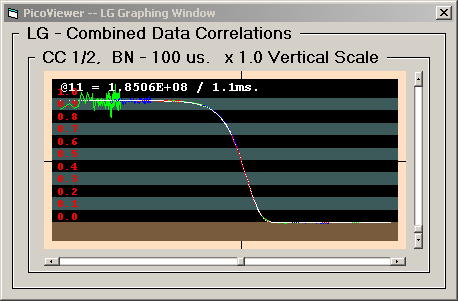
The 'Enable Draw Fit' check box in conjunction with the
selection of a suitable equation and fitting parameters on
the MrqFit -- Data Fitting window allows the
display of a data fit superimposed upon the acquired data.

FC Graphing Window
Clicking the 'FC Graphing Window' menu item opens the
FC Graph:
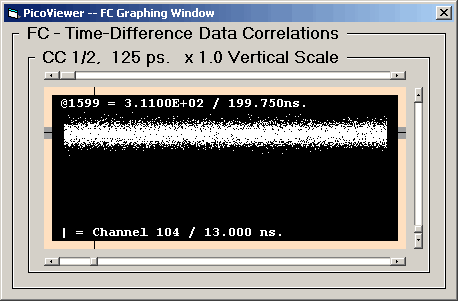
The FC Graph displays a selected 'fast correlation'
spectrum. These spectra were created by the time-difference
correlation process. The valid time-difference range is
131,072 bins at the 125 ps resolution and 262,144 bins
at the 62.5 ps resolution. This results in a valid time range
of 16,384 ns for both resolutions.
The data is scaled so that the channel with the largest
number of counts is displayed at full scale.
The lower horizontal scroll bar allows one to move the
cross hairs (in the tan colored area around the spectrum)
to a particular time. The channel number, counts in the
channel, and the correlation time are displayed at the
top of the spectrum.
The vertical scroll bar may be used to change the
vertical scaling. The current vertical scaling factor
is displayed above the spectrum.
The upper horizontal scroll bar allows one to scroll
the display through the complete data set using the range
specified in the FC Graphing tab. The beginning channel
is displayed at the bottom of the spectrum.
The 'Graphing Control' tab associated with the displayed
FC data is:
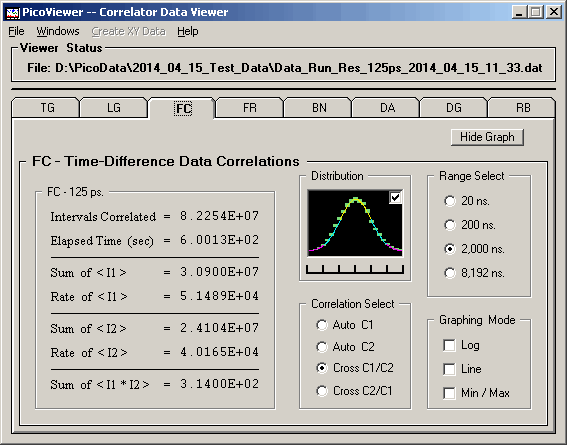
The FC tab options include the ability to select
the time range of the displayed spectrum.
The distribution graph shows a normalized plot of counts per
channel compared to a purely statistical gaussian distribution
expected for the average counts per channel.

FR Graphing Window
Clicking the 'FR Graphing Window' menu item opens the
FR Graph:
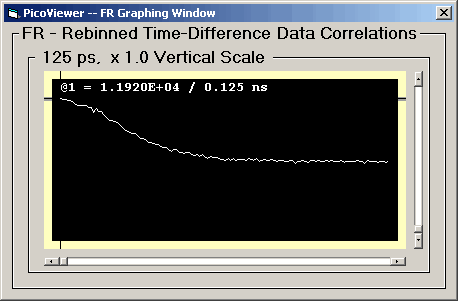
The FR Graph displays a selected rebinned
'fast correlation' spectrum. These spectra were
created by rebinning the time-difference correlation
data into 128 bins of 1, 10, or 100 ns.
The data is scaled so that the channel with the largest
number of counts is displayed at full scale.
The horizontal scroll bar allows one to move the
cross hairs (in the yellow colored area around the spectrum)
to a particular time. The channel number, counts in the
channel, and the correlation time are displayed at the
top of the spectrum.
The vertical scroll bar may be used to change the
vertical scaling. The current vertical scaling factor
is displayed above the spectrum.
The 'Graphing Control' tab associated with the displayed
FR data is:
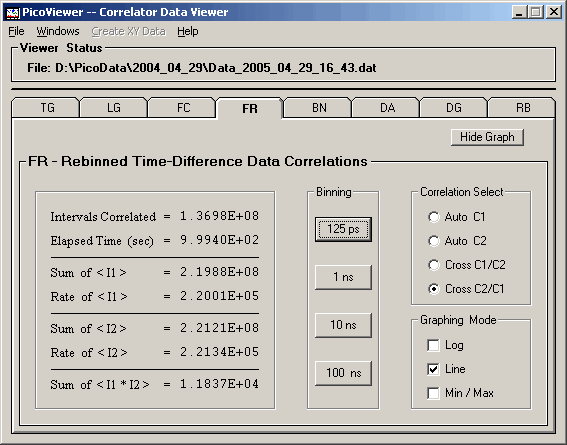
The FR tab options include the ability to select
the time per bin of the displayed spectrum.

BN Graphing Window
Clicking the 'BN Graphing Window' menu item opens the
BN Graph:
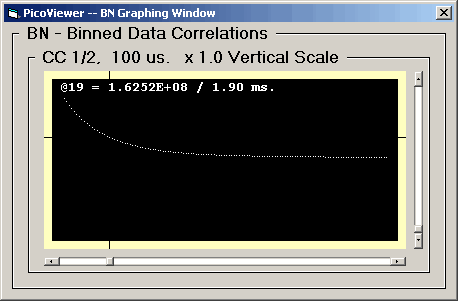
The BN Graph displays a selected correlation
spectrum. These spectra were created by the traditional
processing of counts per bin data. The binned data
correlations were calculated for bin times of 1, 10, 100
microseconds, 1, 10, 100 milliseconds, and 1, 10, 100,
and 1000 seconds.
The data is scaled so that the channel with the largest
number of counts is displayed at full scale.
The horizontal scroll bar allows one to move the
cross hairs (in the yellow colored area around the spectrum)
to a particular time. The channel number, counts in the
channel, and the correlation time are displayed at the
top of the spectrum.
The vertical scroll bar may be used to change the
vertical scaling. The current vertical scaling factor
is displayed above the spectrum.
The 'Graphing Control' tab associated with the displayed
BN data is:
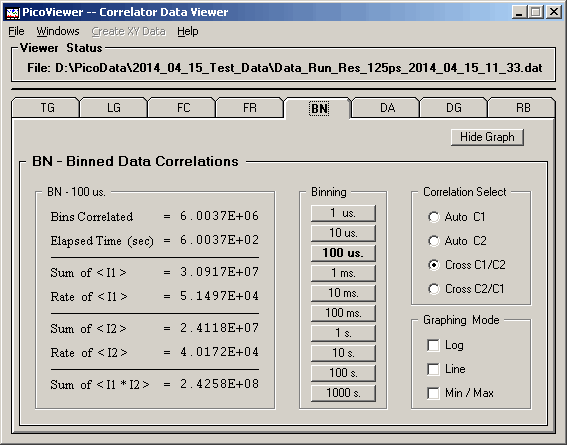
The BN tab options include the ability to select
the time per bin of the displayed spectrum.

DA Graphing Window
Clicking the 'DA Graphing Window' menu item opens the
'Graphing Control' window and the DA Graph.
The DA graphing window is strictly a diagnostic
tool used to study the systematics of the TDC-F1
time-digitizing chip. The TDC-F1 can be programmed
for two resolutions, 62.5 or 125 picoseconds. The
bin times are determined by the propagation of a clock
signal through multiple logic elements. The total
time through some N elements is controlled by an
applied voltage to exactly match some external frequency
source (a precision quartz crystal oscillator). The
locking to the external frequency source (and its phase)
is controlled by an external circuit called a PLL (Phase
Locked Loop).
The manufacture's data sheet explicitly states that
for the high resolution mode there is a strong DNL
(Differential Non-Linearity) between adjacent bins.
However for the low resolution mode there is still a
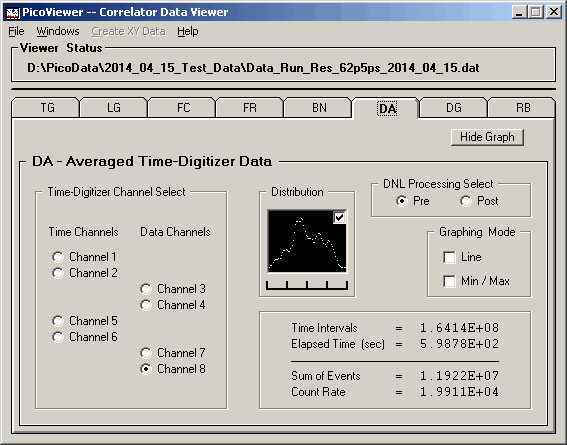
significant DNL component as shown in this first
DA graph with data taken at a 125 picosecond resolution.
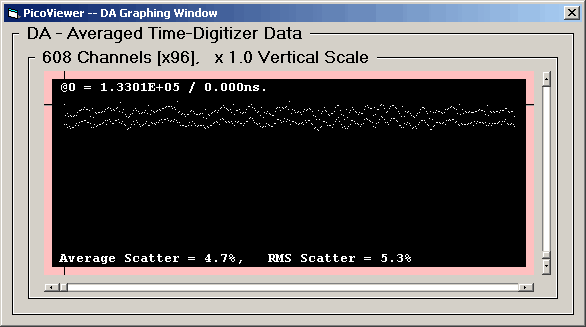
The DA Graph displays a summed spectrum
where the range is 608 bins. This was selected to
be a multiple of the TDC-F1 internal divisor of 152.
The 'x96' signifies that 96 segments of 608 bins,
corresponding to the full scale time-digitizing range
of 58,368 intervals for the Pico Time-Digitizer
Instrument, are summed to provide the spectrum.
The average differential non-linearity is about 10%
between adjacent pairs.
The data is scaled so that the channel with the largest
number of counts is displayed at full scale.
The horizontal scroll bar allows one to move the
cross hairs (in the pink colored area around the spectrum)
to a particular time. The channel number, counts in the
channel, and the channel time are displayed at the
top of the spectrum.
The vertical scroll bar may be used to change the
vertical scaling. The current vertical scaling factor
is displayed above the spectrum.
The average and root-mean-square (RMS) scatter of
data is calculated as an indication of the systematic
error.
The 'Graphing Control' tab associated with the displayed
DA data is:
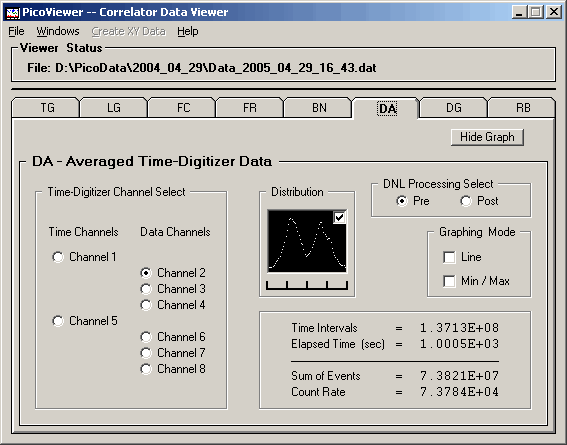
The DA tab also displays a distribution of the
counts in the time bins where the far left is the lowest
counts in a bin and the far right having the largest
counts in a bin. One notes that the distribution of
counts per bin is centered around two dominant times.
The DA graph for the 62.5 picosecond resolution
is very different:
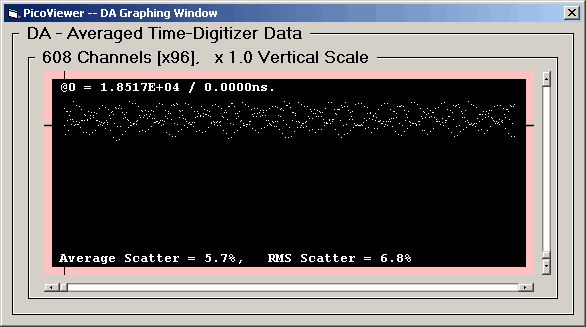
This DA Graph shows a DNL that is predominately
4 channel in character, note how the display appears
to have 4 distinct curves. However the spectrum seems
to repeat every 76 channels. The peak DNL for
this resolution is over 25%.
The DNL appears to have repeating components
with a factor of 2 at 125 ps and factors of 4
and 76 at 62.5 ps. Therefor the DNL correction
algorithm was written as a 4 bin correction repeated
19 times over a 76 bin interval. The 4 bin
correction is suitable for the 125 ps resolution
and the 76 bin correction is suitable for the
62.5 ps resolution. The created algorithm is
used to correct the DNL for both resolutions.
The DNL algorithm is implemented by summing the raw
TDC-F1 time-digitizer channel data modulus 76. The
data is summed for about 15 seconds or until about 4000
counts are acquired in each of the bins, which ever
occurs first. The algorithm then processes the 76 data
bins in 4 bin increments determining how the data in each
bin should be redistributed among itself and the two
adjacent bins to provide an equal number of events in
every channel. The correction fractions are calculated,
saved, and applied during the period required to acquire
the next sample of data. This process continues during
the acquisition cycle allowing the corrections to
dynamically change as required.
Each TDC-F1 time-digitizing channel used for data
acquisition has its own independent DNL correction
array. TDC-F1 time-digitizing channel used for time
markers is not corrected for DNL.
The 'Graphing Control' tab associated with the displayed
DA data is:
The distribution of counts per bin is spread over
a range of times and not localized around a dominant time.
Note the 'DNL Processing Select' option on the DA
graphing control tab. The DA graphs shown so far
are 'Pre' spectra, those with no DNL corrections applied.
The following graphs are for the 'Post' spectra and
have the DNL corrections applied.
First, the 125 picosecond resolution data:
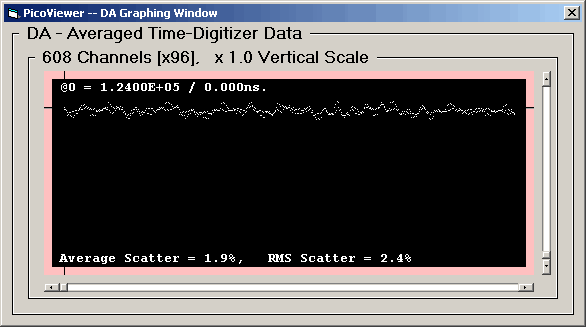
The 'Post' spectra with DNL corrections has removed
the predominate alternate channel differences.
The 'Graphing Control' tab associated with the displayed
DA data is:
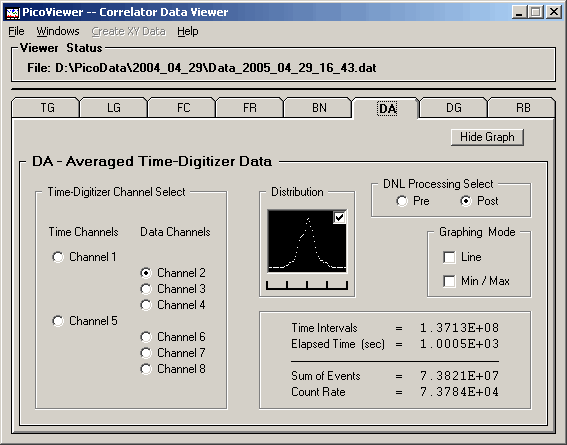
The 'Post' distribution with DNL corrections shows
the central single distribution of counts per channel.
Second, the 62.5 picosecond resolution data:
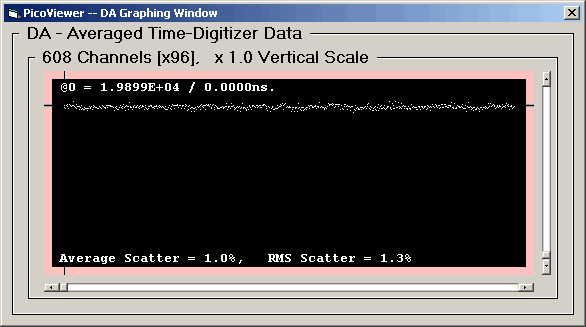
The 'Post' spectra with DNL corrections has removed
the major non-linearity. The average and RMS scatter has
been reduce by a factor of 4.
The 'Graphing Control' tab associated with the displayed
DA data is:
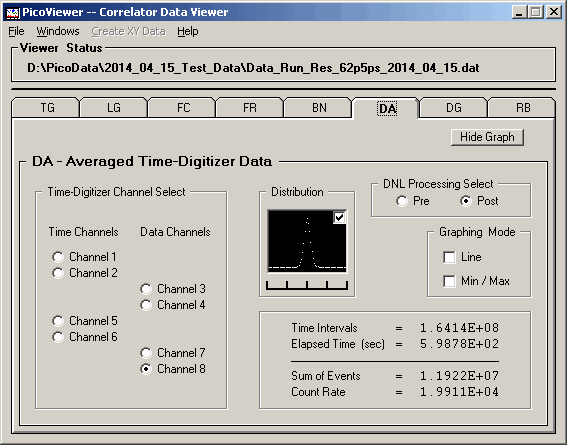
The 'Post' distribution with DNL corrections shows
the the very strongly centralized single distribution of
counts per channel.

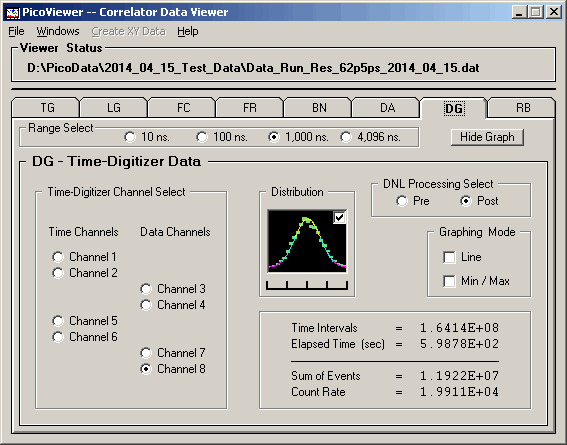
DG Graphing Window
Clicking the 'DG Graphing Window' menu item opens the
'Graphing Control' window and the DG Graph.
The DG graphing window is a diagnostic tool used
to observe the effects of the various corrections
on the data from the TDC-F1 time-digitizing chip.
The 'Pre' and 'Post' correction display options
are available. The TDC-F1 is configured to have a
basic timing conversion interval of 58,368 bins
of 62.5 or 125 picoseconds / bin.
DG (Pre) graph with 125 picosecond resolution data:
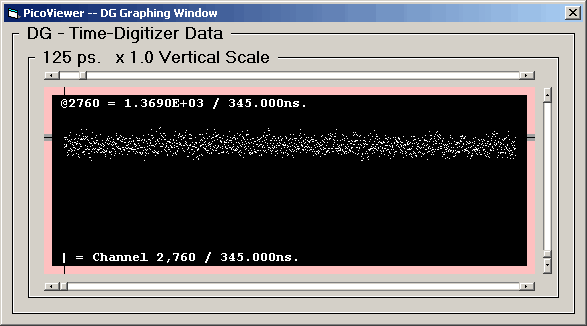
The data is scaled so that the channel with the largest
number of counts is displayed at full scale.
The lower horizontal scroll bar allows one to move the
cross hairs (in the pink colored area around the spectrum)
to a particular bin. The channel number, counts in the
channel, and the bin time are displayed at the top of
the spectrum.
The vertical scroll bar may be used to change the
vertical scaling. The current vertical scaling factor
is displayed above the spectrum.
When the upper horizontal scroll bar is shown one can
scroll the display through the complete data set using
the range specified in the DG Graphing tab. The beginning
channel is displayed at the bottom of the spectrum.
The 'Graphing Control' tab associated with the displayed
DG data is:
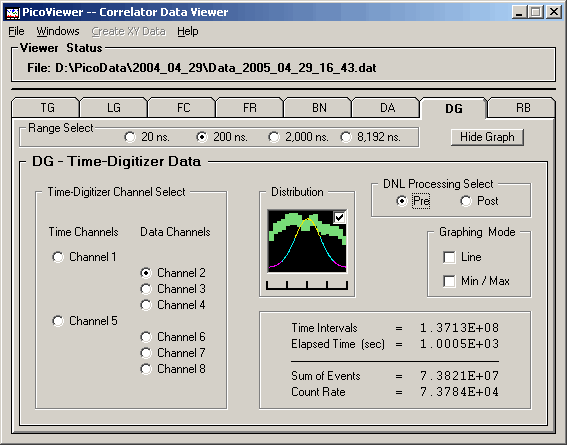
The DG tab options include the ability to select
the time range of the displayed spectrum.
The distribution of counts per bin is spread over
a range indicating it is not statistical in nature.
Note the 'DNL Processing Select' option on the DG
graphing control tab. The DG graphs shown so far
are 'Pre' spectra, those with no DNL corrections applied.
The following graphs are for the 'Post' spectra and
have the DNL corrections applied.
DG (Post) graph with 125 picosecond resolution data:
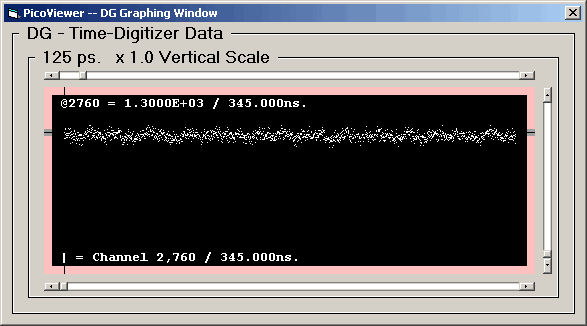
The 'Graphing Control' window associated with the displayed
DG data is:
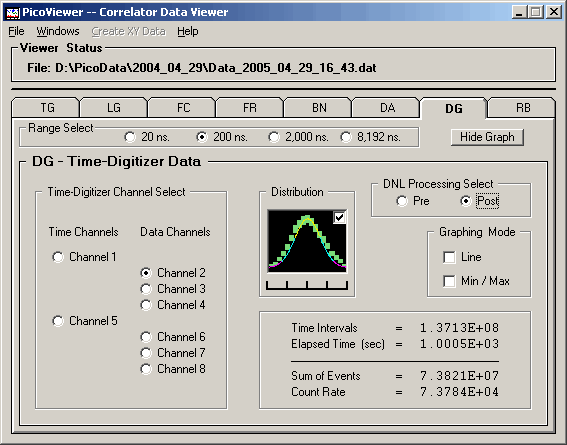
DG (Pre) graph with 62.5 picosecond resolution data:
The 'Graphing Control' tab associated with the displayed
DG data is:
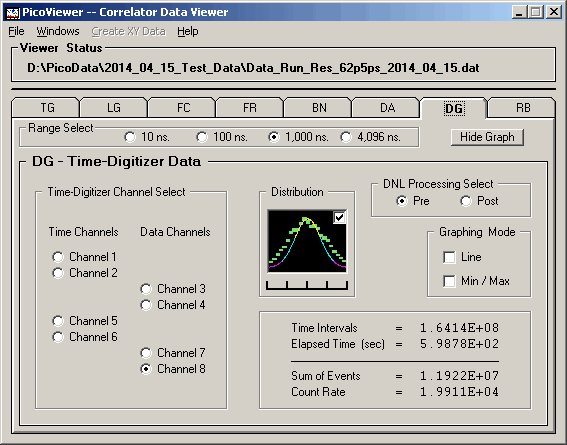
DG (Post) graph with 62.5 picosecond resolution data:
The 'Graphing Control' window associated with the displayed
DG data is:

RB Graphing Window
Clicking the 'RB Graphing Window' menu item opens
the RB Graph:
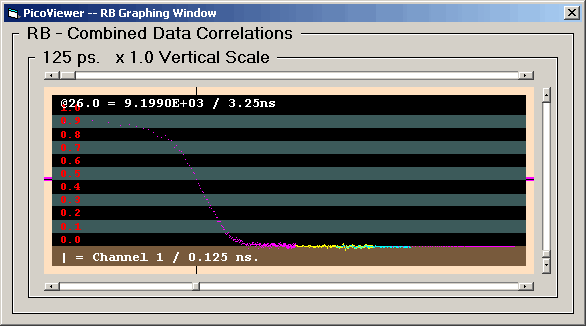
The RB graphing window displays a sequence of rebinned
fast correlation data (FC) which has 131,072 channels or
262,144 channels for resolutions of 125 and 62.5 ps
respectively.
The 'Graphing Control' tab associated with the displayed
RB data is:
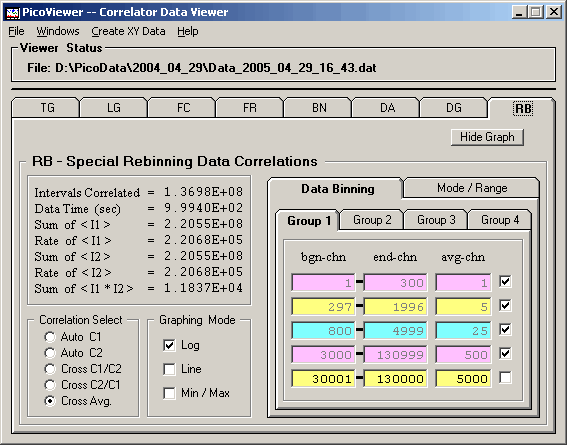
The RB tab options include the ability to select
the correlation spectrum type to be rebinned and the
graphing mode for the displayed data.
The fast correlation data can be rebinned by specifying
the rebinning parameters on the 'Data Binning' tab:
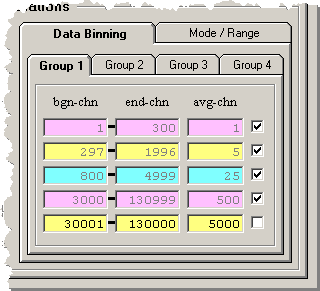
The data rebinning is controlled by the 20 parameter
sets on the four group tabs. The three parameters are
(1) the beginning channel, (2) the ending channel, and
(3) the number of channels to average per displayed point.
To configure a parameter set clear the checkbox on the right,
enter the three parameters, and then set the checkbox to
enable the parameter set. When the checkbox is set the
entered parameters will be checked to verify correct
boundary conditions and updated if required. The data
associated with the checked parameter set will then be
displayed along with all other checked parameter sets.
The 'Mode/Range' tab controls how the parameter sets
are interpreted and the range of the data to be displayed:
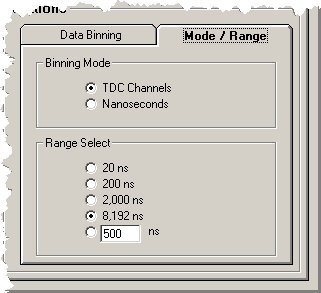
The display mode is selected as channels or nanoseconds.
The display range can be selected from the 4 fixed settings
or a user specified range can be specified and selected.
After specifying and adjusting the parameter sets to
display the data in the form required the data can be exported
to a data file by selecting the 'Create XY Data' item on
the Viewers' main menu:
The output file will be a text file having four
parameters per line corresponding to
(1) the X position in time
(2) the Y amplitude as a normalized correlation spectra
of (counts per bin)/(average counts per bin) with
a value range of 0-2 with a baseline of 1
(3) the statistical error in the Y amplitude
(4) the Y amplitude of any fit function selection, If
a fit was not performed then the output for this
parameter will be 0.
The output data will contain all of the rebinned data plus
all other binned data, FR and BN spectra, in sequential
order excluding lower resolution overlapping data. The
exported data file can be easily imported into an external
analysis or viewing program.

RT Graphing Window
Clicking the 'RT Graphing Window' menu item opens
the RT Graph:
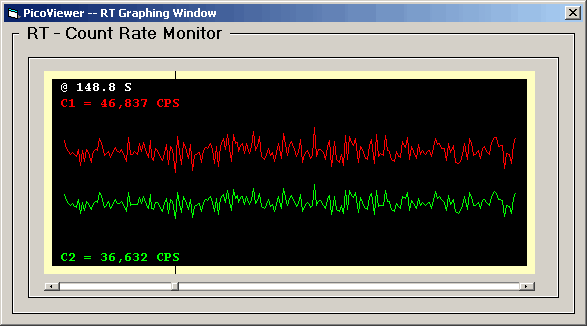
The RT graphing window simply shows the
channel C1 and C2 acquisition count rate as a
function of time.
The horizontal scroll bar allows one to move the
cross hairs (in the yellow colored area around the spectrum)
to a particular time. The channel time and channel
count rates are shown.

MRQ Data Fitting
Clicking the 'MrqFit Fitting Control' menu item opens
the MrqFit Data Fitting Control Panel. The Data Fitting Control
Panel is the control center for configuring the parameters required
to perform data fitting and graphing of the fitting results.
Tab 1: Fitting Configuration
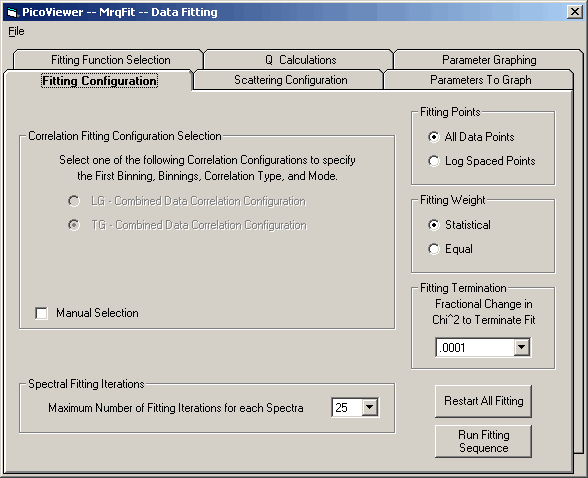
The 'Fitting Configuration' tab contains the following selections:
a) Correlation Fitting Configuration Selection
Automatic configuration selection is the default mode for
specifying the binnings and correlation selection. The
selection is made by
1) Select one of the LG or TG spectra for display.
2) Check the selected 'Enable Draw Fit' box.
A manual selection option is available for specifying the
fitting selection for special circumstances.
The selections are LG and TG. The data fitting is
performed using the spectrum selections from these
graphing windows. The specific detector(s) selected
for display are not relevant to this configuration.
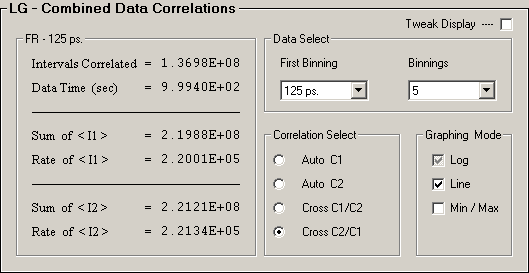
Data Select(First Binning, Binnings) and
Correlation Select(AC or CC) from each detector pair
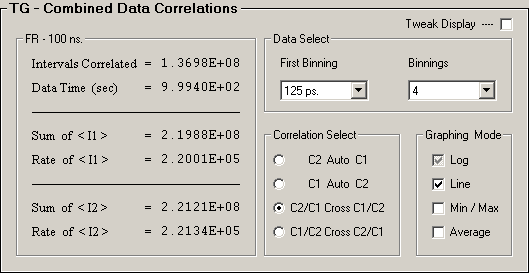
Data Select(First Binning, Binnings),
Correlation Select(AC or CC) from each detector pair, and
Graphing Mode(Average) from each detector pair
b) Spectral Fitting Parameters
All the spectra to fit set in Tab 5 are processed.
Maximum Number of Fitting Iterations for each Spectra [1, 2, 5, 10, 25, 50]
specifies the maximum number of fitting iterations to perform on
a single spectrum at each 'fit' update. The iterations will
terminate if the fractional change limit in Chi^2 occurs.
c) Fitting Points
Select Data Points to Fit:
1) All Data Points - Fitting function uses all
the data points selected by LG or TG options.
2) Log Spaced Points - Fitting function uses data
rebinned into 10 points per decade of time such
that the points are equally spaced on a logarithmic
time scale. The data points are rebinned from all
the data points selected by LG or TG options.
d) Fitting Weight
Select how the fitting function weights the data points:
1) Statistical - The fitting function adjusts the
relative significance of the data point based on
the statistical error of the data. Data with
smaller statistical errors have more significance.
2) Equal - The fitting function treats every data
point with equal significance. Statistical
errors are not used.
e) Fractional Change in Chi^2 to Terminate Fit
This parameter specifies the lower limit of the absolute fractional change
in the normalized Chi^2 value to terminate the fitting iterations. The
selections are [.01, .001, .0001, .00001, .000001]. The default is .0001.
The two comand buttons:
'Restart All Fitting' resets the fitting functions to their initial
values and prepares for a new fitting sequence.
'Run Fitting Sequence' initiates a fitting sequence.
Tab 2: Fitting Function Selection
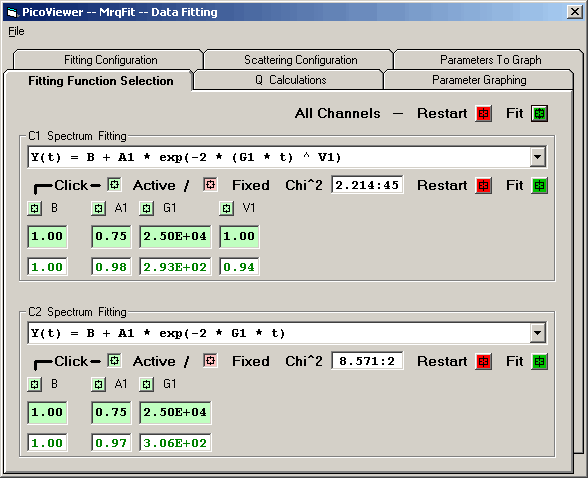
The loaded data is processed using the Levenberg-Marquardt
method attempting to reduce the value Chi^2 of a fit between a
set of data points x[1..ndata], y[1..ndata] with individual
standard deviations sig[1..ndata], and a nonlinear function
dependent on n coefficients a[1..n]. The algorithm allows
selected initial parameter values to be held constant during
the fitting process.
The All Channels Restart and Fit buttons respectively:

Restart -- Resets the fitting functions to their initial
values and prepares for a new fitting sequence.
Fit -- initiates a fitting sequence.
The 'Fitting Function Selection' tab allows the selection of a
fitting equation for each of the 2 detectors. The equation is
selected from the dropdown box:
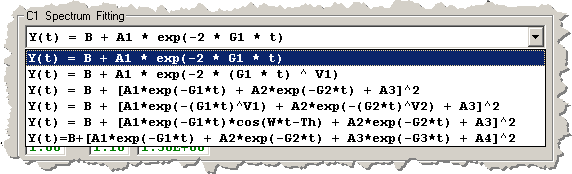
The equations and corresponding displays are:
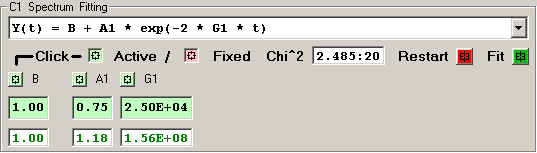
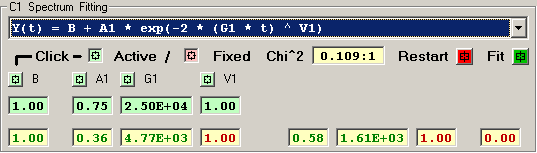
1) Single Exponential [3 independent terms]
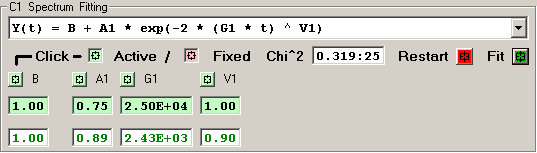
2) Single Stretched Exponential [4 independent terms]
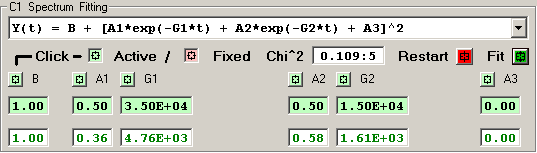
3) Double Exponential [6 independent terms]
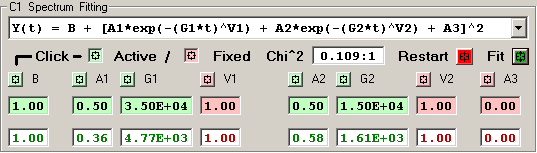
4) Double Stretched Exponential [8 independent terms]
5) Double Exponential with Oscillatory Term [7 independent terms]
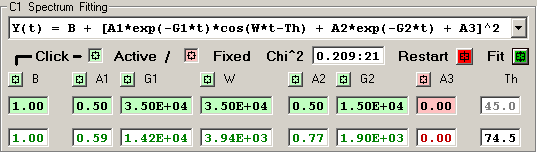
6) Triple Exponential [8 independent terms]
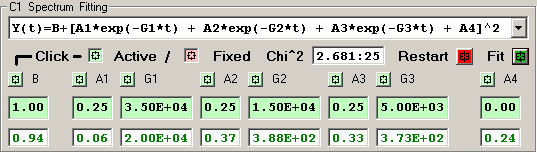
The features of the fitting function display are described
in detail for each row of elements:

The Chi^2 panel displays the normalized Chi Square value
followed by the number of fit iterations before the fitting
was terminated by the Chi^2 limit or iteration limit.
Pressing the RED Restart button initializes the fitting
process for this detector by clearing the current fit
values and loading the initial parameter values for the
fitting function.
Pressing the GREEN Fit button initiates a fitting
sequence for this detector. The number of iterations
is determined by the values selected in the Maximum
Number of Fitting Iterations on Tab 1.

Each independent fitting parameter has an associated
button which specifies if this parameter is a variable
or fixed value. A Light-Green button indicates a varible
parameter and a Light-Red button indicates a fixed value.
Pressing a button toggles the parameter from variable to
fixed or fixed to variable. Changes to the parameter
type become effective when the RED Restart button is
pressed or the 'Restart All Fitting' button is pressed
on Tab 1.

The initial fitting function parameter values are
shown with a Light-Green background for variable parameters
and Light-Red for fixed parameters. New values for these
parameters may be entered into the boxes, if an invalid
value is entered the original value is restored. Changes
to the parameter values become effective when the RED Restart
button is pressed or the 'Restart All Fitting' button is
pressed on Tab 1.

The resulting fit values are displayed as GREEN for
variables and RED for fixed parameters.
An initial fitting function parameter value may be
loaded from the result fit value by 'double-clicking'
the result fit value.
Note:
If the fitting function selection is changed the new
fitting function initial parameters will be displayed.
However, the previous fitting function is still active
and the displayed fit values will continue to be updated.
This condition is indicated by the Light-Yellow background
for displayed fitting results. Changes to the fitting
function become effective when the RED Restart button is
pressed or the 'Restart All Fitting' button is pressed
on Tab 1.
The 'MrqFit Data Fitting' File Menu:

gives the option to save the fit values to a file.
A standard Save As file dialog box will open:
Tab 3: Scattering Configuration
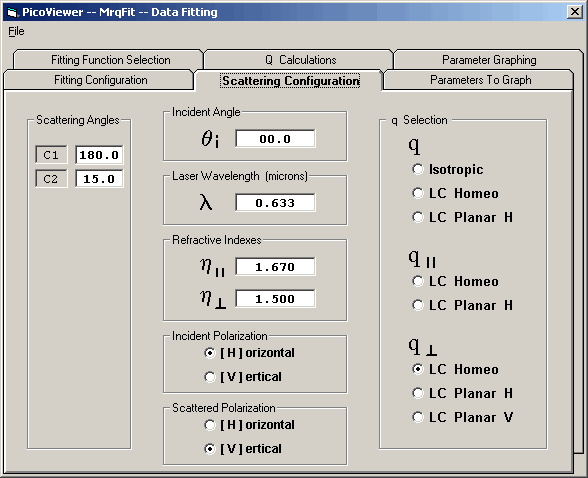
The scattering configuration tab is used to specify the
scattering parameters and configuration. These parameters
will then be used to calculate the 'q' values of the scattered
photons. The parameters and configuration items are:
1) Detector scattering angles in degrees
2) Laser light incident angle in degrees
3) Laser light wavelength in microns
4) Parallel index of refraction
5) Perpendicular index of refraction
6) Incident Polarization ( [H]orizontal or [V]ertical )
7) Scattering Polarization ( [H]orizontal or [V]ertical )
8) Sample Type Selection:
a) q
1) Isotropic
2) LC - Liquid Crystal Homeotropic
3) LC - Liquid Crystal Planar [H]orizontal
b) q - parallel
1) LC - Liquid Crystal Homeotropic
2) LC - Liquid Crystal Planar [H]orizontal
c) q - perpendicular
1) LC - Liquid Crystal Homeotropic
2) LC - Liquid Crystal Planar [H]orizontal
2) LC - Liquid Crystal Planar [V]ertical
The square of the 'Q Selection' values may then be used as the
Y-Axis scaling of the 'Parameter Graph' displayed in Tab 6.
Tab 4: Q Calculations
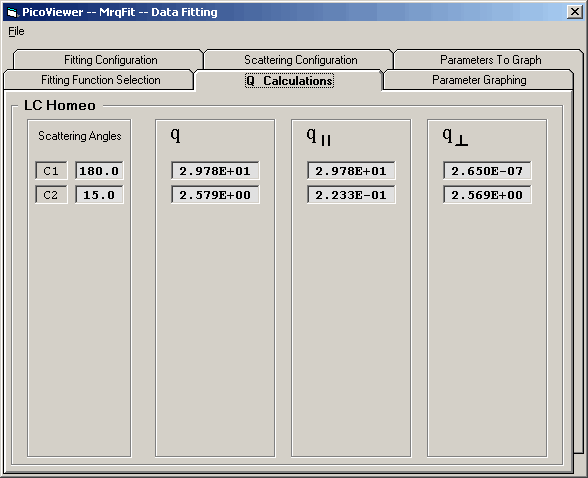
The Q Caculation Tab simply shows the calculated values for
q, q-parallel, and q-perpendicular for each detector scattering
angle based upon the parameter values and q selection in Tab 3.
Tab 5: Fit Parameters to Graph
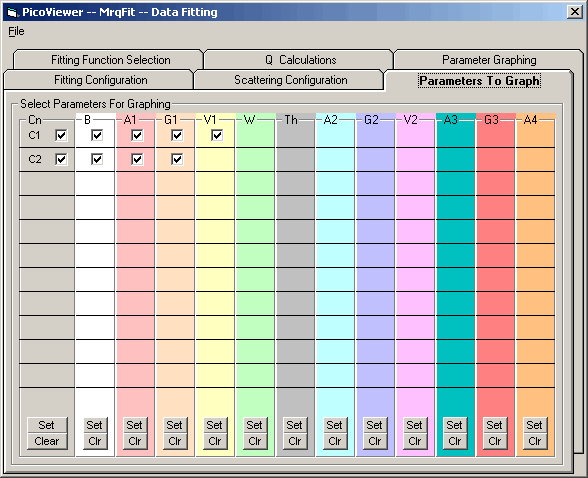
The 'Fit Parameters to Graph' tab enables fitting of selected
detector data (C1 - C2) and the graphing of selected parameters
during data acquisition.
The detector (C1-C2) check box(s) must be checked to enable
data fitting for a specific detector. A detector's parameter
check box(s) must be checked to enable graphing of the parameter
in the 'Fit Parameter Graph' in Tab 6.
Note:
The parameter check box selections for detector (C1-C2) are
associated with the currently selected fitting function.
Changing the fitting function for a detector will change the
parameter check box selections.
Note:
Graphing windows LG or TG DO NOT require a detector check box
to be checked. The 'Enable Draw Fit' check boxs on the LG and
TG 'Graphing Control Window' tabs independently enable
fitting for the spectra specified in the Graphing Control Window.
Tab 6: Fit Parameter Graphing
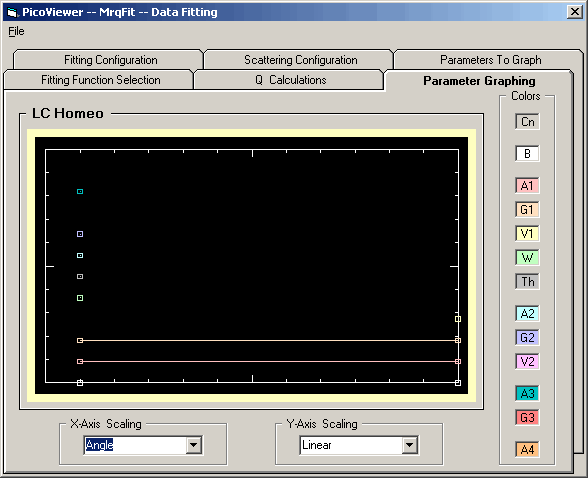
The 'Fit Parameter Graphing' tab simply displays the selected
parameters from the data fitting. The parameter data is color
coded as noted.
The X-Axis drop down selection box
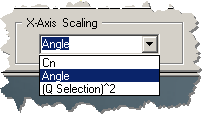
selects the X-Axis scaling as either the detector number,
detector angle, or the square of the 'Q Selection' value
from the 'Scattering Configuration' of Tab 3.
The Y-Axis drop down selection box
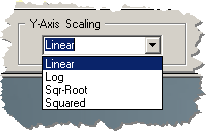
selects the Y-Axis scaling as one of the following:
1) Linear
2) Log
3) Sqr-Root
4) Squared
The X values are typically normalized to display at .5 or
.25 and .75 for 1 or 2 detectors, from 0 to 180 degrees for
angles, and 0 to 1, 2, or 5 times a power of 10 for Q^2 values.
The Y values for parameters B, A1, V1, A2, V2, A3, and A4
are displayed with a range of 0 to 1. The Y values for
parameters G1, W, Th, G2, and G3 are normalized to display
from 0 to the maximum value of each parameter.



























